Use case 5: Ranking interactions driven by short linear motifs
Problem
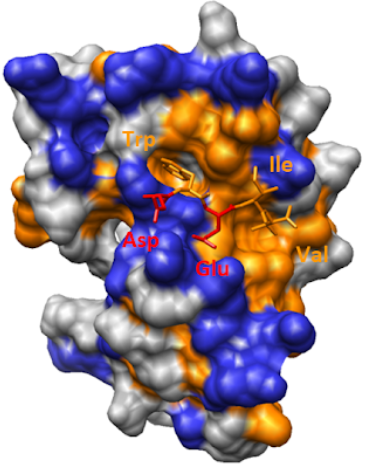
We would like to predict whether AlphaFold2 scores can be used to rank short linear motif based interactions. We have an experimental setup that use different peptides as bait and use protein domains as prey. We would like to compare assay results with computational predictions. Using BLAST and peptide search we prepared a list of UniProt ACs, containing domains and peptides. We use Colabfold with default settings to predict combinations of different interactions. We do not use template structures.
Initial dataset
We have a list of UniProt identifiers, some of them were shown to bind to each other.
?
We run AlphaFold-multimer without templates and compare it with our experimental setup. Can we do that?
!
Use full length protein set with the AlphaFold-multimer training date as cut off (01.10.2021.)
Proposed protocol with BETA
- ColabFold with default settings uses AlphaFold2-Multimer. According to the README provided to AlphaFold the model for the multimer version was retrained on 30.09.2021.
- Go to the BETA download page and select the benchmark version. For the benchmark set we use full length proteins. For date we select the closest date after the cutoff, which is 01.10.2021.
- The intersection of the original benchmark set (step 1) and the filtered set from BETA (step 2) should be used to assess predictions.
- (Alternatively interacting full length proteins can be also selected to compare results with interactions stored in BIOGRID)